Analyzing postprandial metabolomics data using multiway models: A simulation study
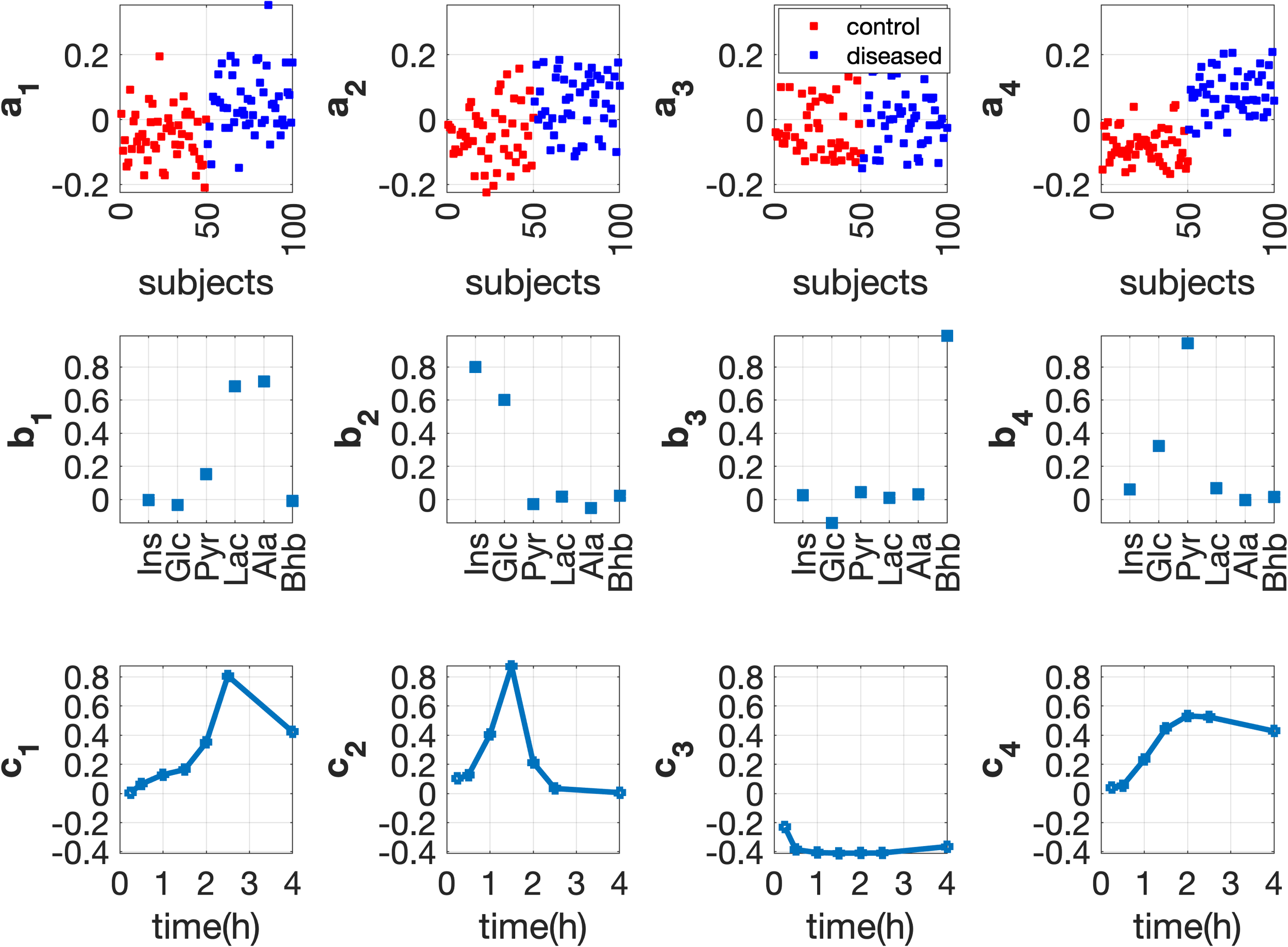
Patterns extracted by a 4-component CP model from a subjects by metabolites by time array.
How similar are we in terms of our metabolic response to a meal? Can we stratify people according to their metabolic response, understand differences and similarities among groups of individuals and ultimately capture early signs of diseases?
Answering these questions requires the analysis of time-resolved postprandial metabolomics data. Postprandial metabolomics measurements at several time points from multiple subjects are in the form of a three-way array: subjects by metabolites by time points array. Traditional analysis methods are limited in terms of revealing subject groups, related metabolites, and temporal patterns simultaneously from such three-way data.
In our recent study, we have introduced an unsupervised data analysis approach based on the CANDECOMP/PARAFAC (CP) model for improved analysis of postprandial metabolomics data guided by a simulation study. Because of the lack of ground truth in real data, we generate simulated data using a comprehensive human metabolic model. This allows us to assess the performance of CP models in terms of revealing subject groups and underlying metabolic processes. We study three analysis approaches: analysis of fasting-state data using Principal Component Analysis, T0-corrected data (i.e., data corrected by subtracting fasting-state data) using a CP model and full-dynamic (i.e., full postprandial) data using CP. Through extensive simulations, we demonstrate that CP models capture meaningful and stable patterns from simulated meal challenge data, revealing underlying mechanisms and differences between diseased vs. healthy groups.
This study has been carried out together with our collaborators at the University of Groningen, University of Amsterdam, University of Copenhagen and COPSAC - special thanks to our postdoc Lu Li for tirelessly working to put together such a comprehensive study!
The preprint is now available on bioRxiv:
L. Li, S. Yan, B. M. Bakker, H. Hoefsloot, B. Chawes, D. Horner, M. A. Rasmussen, A. K. Smilde, E. Acar. Analyzing postprandial metabolomics data using multiway models: A simulation study, 2022